I frequently need to store large binary data like parsed text objects, trained ML models, or custom data types as part of my data analysis workflow. For me, SQLite and other database engines are obvious choices because I ususally need to record metadata like algorithm hyperparameters, evaluation results, or timestamps and any other information that might be relevant when I need to look back at various models I have run. Using relational databases to store large binary data is typically ill-advised, so my solution was to implement a custom column type that handles insertion of binary data by saving to the filesystem and including only the file path in the original database.
If you’ve been attempting to keep track of multiple models generated from your analyses without databases, you’ve likely been generating filenames like `modelA_a=0.1_tau=3_iter=5_runs=100.pic’, which include information about the stored model with some metadata. The metadata is hard to sort through because you will be reading raw filenames or writing parsers to automatically scan the files to find a model you are interested in. In contrast, traditional databases are great at storing this type of metadata, but innapropriate for storing large binary data alongside it. The implementation of my solution in doctable allows us to treat these columns just as if they were regular binary (or text) data being stored in the database directly, while actually storing the data in the filesystem for faster read and write times.
In case you are not familiar, doctable is designed to
provide an object-oriented interface for working with relational
database tables. It is database engine independent because it is built
on SQLAlchemy Core, but does not provide object-relational mapping (ORM)
functionality - it simply works as a way to describe database schemas in
Python code, generate queries, and work with query results through a
Python interface. I used SQLAlchemy’s custom data type feature to
implement my solution to the binary storage problem, so it can be
accessed just as if it were any other data type.
Implementation
From the doctable perspective, these special file columns look like
any other column used to stored pickled binary data. I created a few
examples that might be helpful for understanding the main interface
implemented in doctable here.
I implemented this feature by subclassing
sqlalchemy.types.TypeDecorator to a custom
PickleFileType class and overloading the
process_bind_param and process_result_value to
implement the insert/update and select functionality as you would any
other custom sqlalchemy type.
Each time a query is executed, sqlalchemy constructs a new
PickleFileType and is passed a reference to a persistant
FileTypeControl object which was created when the
DocTable was instantiated. Each file-based column maintains
a FileTypeControl object to maintain state between queries
and bridge messages to and from the DocTable. When an
object is inserted into the database, the PickleFileType
object creates a random filename for the payload data, and writes the
pickled object to disk. It then writes the filename directly into the
SQLTable to keep track of the associated file. When an object is
selected from the database, the PickleFileType retreives
the filename and reads/unpickles the file into the returned object. From
the interface, insert/update/delete operations appear as if you were
inserting or retrieving data in the database directly.
Benchmark results
I set up a benchmark where I created two different databases: one that includes a payload column where the data is to be inserted directly into the database, and another that includes a payload column where that stores the data into a file in the filesystem and inserts only a filename into the actual database.
The benchmark consists of three different measures. The first is a measure of insert time: how long it takes to store a set of binary objects into the database. The second is a measure of how long it takes to select the index column of the database. The hypothesis behind this measure is that inserting large quantities of data into the table will create larger distances (in terms of disk addresses) between each successive row, and thus slow down select statements even when the payload column is not included in the query. The last measure is the query for the actual payload, which is intended to test the raw read speed.
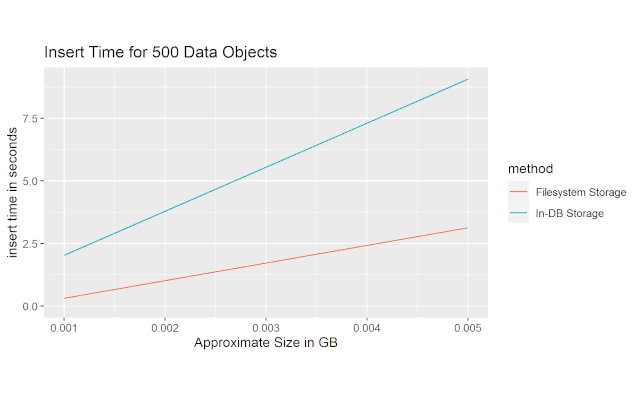
This first figure shows the average ammount of time it took to insert 500 objects into an SQLite database on my machine at various data payload sizes. We can see that the file-based solution I implemented shows a ~3x improvement over inserting directly into the database.
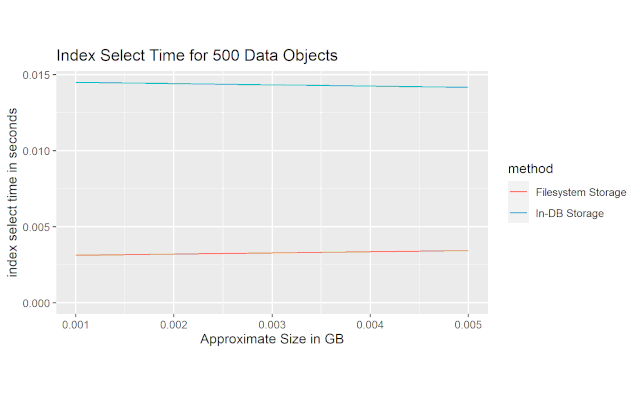
This next figure shows the average ammount of time it took to select just the index column of the table. We can see that the file-based solution is approximately 5x faster than the in-database storage solution.
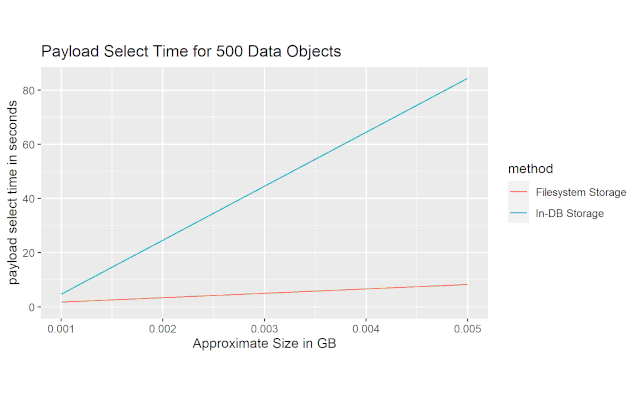
This last figure shows the time it took to load the actual data payload. The file-based solution is much faster than the in-database storage solution, and the multiplier appears to increase with filesize.
Conclusion
From these results we can see that the file storage-based solution improves performance significantly according to these three measures. Of course, it is difficult to project these results into extreme cases where we are storing extremely large data payloads or a ton of really small payloads, but I think it does provide insight into some reasonable cases.