I wanted to reflect on a few design principles for software design of your data science pipeline that I have come to after doing data analysis for research and consulting over the last 7 years. Some of these projects were quick and simple, while others started out that way and quickly became cumbersome. It is the latter category of projects that encouraged me to propose these principles. While some are derived directly from accepted software design principles, others directly contradict them - in these cases, I relied on my own experience on the patterns we fall into and the needs of data science projects to propose alternative approaches. First I list the principles, then I go into more details.
-
Know thy data: The structure of data should be explicit in your object design. Use objects to explicitly represent your source or derivative data, avoid dataframes or nested iterables when possible, and implement data validation upon ingestion. This builds upon the Zen of Python principles suggesting that "explicit is better than implicit" and "flat is better than nested." More generally, building explicit structure into your data pipeline can reduce the potential for errors and provide some built-in gaurantees about available data.
-
Make object pipelines obvious: Go nuts with factory method construtors. Place the logic for constructing an object into a factory method instead of the default constructor. This follows naturally from the Zen of Python principles "in the face of ambiguity, refuse the temptation to guess" and "there should be one-- and preferably only one --obvious way to do it." This creational design pattern from Gang of Four is particularly useful in data science scenarios where classes are often data representations. Creating separate factory methods for each type of input data can make dependencies clearer and make objects easier to test and extend.
-
Validate upstream when possible, and anywhere when not. Create gaurantees for your data by adding validation code when you are able - ideally as far upstream in your data pipeline as possible. This validation code should ideally be used in addition to unit testsing, but any validation is better than none. I will show some examples that use this principle later.
-
Handle missing data downstream.
Not all downstream analyses will be affected by an original
Custom exceptions create a lot of flexibility when working with missing data.
-
Version almost everything. Version both your code and data files/databases. While this can (and will) lead to a high level of code rot, it will be critical for keeping track of which pipelines and settings were used to generate each intermediary or final piece of data. This is especially important when you try different parameter sets and need to know exactly which parameters were used to generate each result. Git is the bare minimum here, although using the releases features there could be a good temporary substitute. While following this in every situation might be overkill, I recommend doing it for especially critical parts of your code - particularly for your final (or communicated) result result files.
-
Create param objects files that live in your code. Create explicit param objects to store parameters used to generate each intermediary or final dataset in your project. This follows directly from the previous two points, and it will make it easy to identify which parameters were used to create each set of files. I have found this to be a more common issue than I expected.
-
Make I/O explicit in your top-level scripts. Read and write functions should always appear in your toplevel script (e.g. main file or toplevel script) and it should be clear which type of data you are reading/writing. By this principle I mean that it should be easy to tell which types of data are being ingested and which types are being saved through a quick scan of your script.
Theory of Data Analysis Pipelines
It will be helpful to first outline a skeleton of a data science project to see why some design patterns, programming principles, and project management strategies are better suited for data analysis than others. By definition, data analysis involves the transformation of one type of data to another. For instance, maybe you are given a csv file and asked to generate a figure, or retrieve json data from an API and asked to produce a regression table. In these instances, your clients (whether it be journal reviewers, customers, or managers) expect you to transform some type of input data into data of some format for human interpretation.
One way to think of these projects is as a pipeline: the pipeline starts with your source data and involves a series of transformations produced until the final result is produced. The figure below shows an example data pipeline. Each block represents some kind of structure that your data takes - essentially a set of measurements and relationships between them - and lines represent the algorithm you use to transform it. Some of these structures may be written to disk and loaded later down the pipeline in another script, and some of the transformations may appear as part of every script. Importantly, these datasets have dependency relations such that they are all derived from either the client source data or some outside source, such as a fitted statistical model or a sentiment analysis dictionary used to transform your client source data.
As an analyst, your job is to build data pipelines that minimize the likelihood of errors, maximize reproducability (by others and yourself), and optimize flexibility for future changes. The following principles are designed to support these goals.
1. Know thy data: The structure of data should be explicit in your object design
By this I mean that your data pipeline should keep track of the structure of your source and intermediary data without being presented with the input data itself. While it is tempting to pass dataframes (often from csv files) or nested iterables (e.g. lists of dictionaries) through your data pipeline because they are robust and you can use them to do many things, these exact attributes serve to increase the risk of errors in your code and make it harder for the programmer to read or modify in the future. Adding more structure to your design can provide built-in gaurantees about the structure of your data at any point in the pipeline, even in the absence of validation procedures.
At first glance it seems like building in more structure is tedious and unnessecary, but we've all experienced the scenario where very simple analyses become increasingly complex as our project scope widens, we need to make changes based on intermediary results, or the objectives of the project change entirely. In the real world, simple projects inevitably become more complicated over time, and building more explicit structure into your pipeline designs can help to mitigate some of the issues that arise in that case.
Because data is a noun, for most languages I recommend creating immutable (unchangable) objects with a fixed set of properties to represent collections of data. On a theoretical level, let us first imagine that our source data includes various measurements taken on a set of plants. Assuming there is overlap between the measurements taken on each plant and the number of measurements can be enumerated and is no greater than 30 or 40, I recommend creating an object to represent each plant, and parsing input data into a sequence of those objects.
In this case, the first step of your data pipeline should be to convert your input data into a sequence of these plant objects. By placing the data into these objects, you are committing to some gaurantees that can be taken for granted by any function that attempts to use these objects. First of all, we gaurantee that every plant will have the same set of properties that we can work with (whether they represent missing data or accessing them raises an error is a different question). Without running your code, you can detect
Any static analysis assistant in your IDE like Intellisense etc will recognize cases when you try to access an attribute that does not exist.
when passing this data to a future function. Any function this data is passed into
know that every plant will have the same set of properties. If
with enumerated properties to represent a piece of data. In the examples below
They often encourage you to examine data through introspection at intermediary points in your pipeline, and make it difficult for code validators to keep track of the structure of that data at each point.
In data science it is particularly important to be able to track down the procedures used to produce a given data result, and building more explicit structure into that pipeline can make it easier to understand and change later.
For illustration purposes, I'm going to use the iris dataset loaded from seaborn. The dataset is provided as a single dataframe with five columns (of which we will use the 3 shown here).
import seaborn
import pandas as pd
iris_df = seaborn.load_dataset("iris")
iris_df = iris_df[['sepal_length', 'sepal_width', 'species']]
iris_df.head()
The first five rows look like the following:
sepal_length sepal_width species
0 5.1 3.5 setosa
1 4.9 3.0 setosa
2 4.7 3.2 setosa
3 4.6 3.1 setosa
4 5.0 3.6 setosa
Now I will illustrate potential issues
The problem with implicit data structures
For illustration, I created a simpler diagram with two linear data pipelines depicting the transformation of the input data into an intermediate data structure which is changed into the final data to be shared with the customer (a table or figure, let's say). As I noted earlier, almost every part of your data analysis pipeline will look something like this. In the top example, we do not keep track of the structure of the input or intermediate data in our code explicitly, wheras in the bottom pipeline we represent them as objects A, B, and C. The idea is that pipelines with explicit references to data structure in the code make it easier to understand what each transformation is doing - in theory, we (and the static analyzer in your IDE) could understand the entire pipeline without ever running our code.
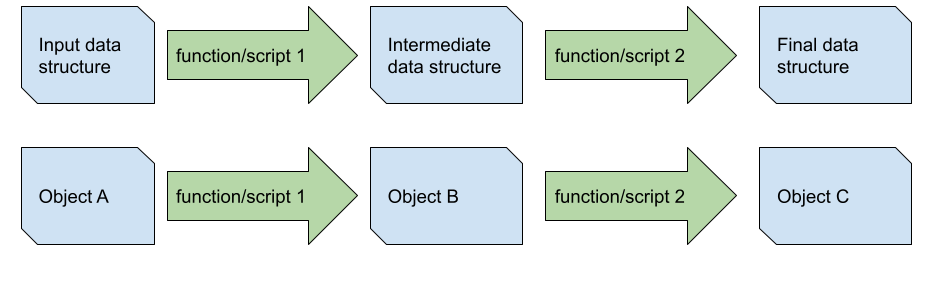
As a hypothetical, let's say you are seeing a potential issue in your final data structure - a figure, let's say - and you want to investigate why you observe a given value. First you hypothesize that the issue may have been with function/script 2, and so we first need to understand the structure of the intermediary data which it transformed. There are three approaches to understanding the intermediate data structure when we have not been explicit in our code:
- remember the structure of that data - generally a terrible idea in software design because you may be looking at this years later or someone else may be looking at it;
- run the first pipeline component and use some runtime introspection tool (breakpoints, print statements, debuggers, etc) to look at the data - possible but clunky and time-consuming; or
- do some mental bookkeeping to trace the original input data (which may also require introspection) through the pipeline - also a time-consuming activity. If, however, you had built explicit object definitions into your code, you would know the structure of the data exactly without looking at the code used to generate it. Thus, it separates the logic of your operations from the structure of your data.
Instead, I recommend creating objects to represent atomic or higher-order pieces of data that you ingest as a starting point. As an exampl, use
For example, if you read in a csv file as a dataframe, consider creating a class definition that represents a single row of that dataframe and include the code to parse that data within the same class. While json data may be more heirarchical, there are almost always equivalent data units at various levels - you can create a data structure for each level. Using this approach, you can know the structure of your data at any point in your pipeline and your IDE's static analyzer can identify any downstream issues that arise from a change in that data structure. This can be an invaluable tool for
Case Against Dataframes
While dataframes are important data structures that a large suite of languages and packages have been built around, I have two primary concerns about using them as a central feature of your data pipelines: (1) all of the problems we observe above, and (2) they are often the wrong tools for the job (performance-wise) - even though they may be fine for many tasks involving small datasets.
The first point appears to be acceptable for many data scientists given that it is common to use Jupyter or R Markdown notebooks to write large portions of code. Except in initial development or in your toplevel scripts, I recommend using project file structures that are recommended for your language of choice - in Python, this means separating functions and classes (including the data containers) into modules, but there are equivalent recommended project structures for most langauges. Data science projects in particular tend to grow in scope or change in structure often, so modular project structures are especially important. The more complex your code becomes, the more important this is.
More concretely, lets refer to the iris example dataset we loaded. We access a column of that data using a subscript or as a property of the dataframe (although be careful with the latter):
iris_df['species']
iris_df.species
Or, similarly in R:
iris_df[:,'species']
iris_df$species
The problem with this is that you have no gaurantees that this property exists with this name in your input data. Even though the R and Python versions are both written as if the columns are object properties, they are not - they are simply syntactic sugar used to make it feel like they are - the reader, and your static analyzer, cannot gaurantee they exist except in runtime.
Sure, you could run a verification or transformation function that selects/orders columns and does some validation, but this code is implemented as part of the script loading the data, not in the definition of the data itself.
iris_df = iris_df[['sepal_length', 'sepal_width', 'species']]
assert((iris_df['sepal_length']>0).sum() == iris_df.shape[0])
In short, you have no gaurantees that a property will exist in runtime (assuming, as in the case of weakly typed languages, you follow the hints/gaurantees that you yourself provided).
I a also caution against using dataframes for performance reasons. Traditional data structures like sets, dicts (any kind of map), or lists (arrays) in Python all have implementations that are akin to different tasks. If you want to store an unordered set of unique elements, use a set object. If you want to perform lookups from a string or other hashable object to another object directly (i.e. in O(1) time), use a dict. If you want to store an ordered sequence of elements that can be indexed by their order in the sequence, use a list.
As an example of this issue, lets say that you want to perform a join on a two unindexed dataframes A with n rows and B with m rows that have a many-to-one relationship: that is there are zero or more rows of A that correspond to a single row of B. Because dataframe columns are implemented as sequential elements, the time it will take for this join to complete will be proportional to n*m: you would iterate through every element of A to find the associated row in B (note that this is worst case: O(n*m)). This is the wrong tool for the job because it could be done faster with a dictionary or hash map that allows you to look up the associated row in dataframe B instantly, so the time would be proportional to n (even if you have to create the dictionary first, you end up with n + m ~ O(n)).
Note that indexing dataframes is an operation that would allow for faster joins (typically through binary search, which is O(n*log(m))), indexing and multiindexing implementations are somewhat clunky to use and often involve explicit indexing between transformations. It is worth noting they exist, though I don't often see them used.
Dataframes can also be more memory intensive because joins and most other operations often create copies of data - even when it may be unnessecary. Be wary of this as your dataset gets larger.
As an alternative, I recommend creating an object to represent each row of your dataset, and parsing each row using a factory method - I will give some examples later.
Nested Iterables are Also Bad
It may also be tempting to use raw nested iterables like sets, dicts, or lists either because they follow directly from the structure of the input data (especially json data) or because they solve the second issue I have with dataframes - you can use the right tool for the job. My main concern with these structures is that they can get very complicated with high levels of nesting and requre missing data/error handling at every point of usage.
As another example, let us consider a json dataset parsed as a list of dictionaries with properties associated with irises. The type hint for the parsed structure would be typing.List[typing.Dict[str, typing.Union[float, str]]].
[
{
"sepal_length": 5.1,
"sepal_width": 3.5,
"species": "setosa"
},
{
"sepal_length": 4.9,
"sepal_width": 3.0,
"species": "setosa"
},
...
]
Sqy we want to get the average petal length for irises in our dataset, so we do this:
import statistics
statistics.mean([iris['petal_length'] for iris in irises])
The problem here is the same as that of selecting columns in dataframes: we have no gaurantees that each iris will include a member called 'petal_length' until runtime. We can't know it exists unless we recall the data being passed in, and static analysis tools cannot help us.
The solution to these problems would again be to create a class representing a single Iris object and parsing them into a list of these objects. Even better, we could encapsulate the sequence of irises to add additional convenience. I will show some examples in the next sections.
How to make explicit data structures
Creating an explicit object definition for your data will provide built-in validation because you are making gaurantees for what your data will look like and how you will access its attributes downstream. Consistent with broader motivations for using OOP, you could build out your entire data pipeline beginning-to-end with just the knowledge of the properties of the data you will be using - it can be agnostic to the structure of the data you take as input. You could even build downstream components of your pipeline before writing upstream - an approach that requires dicipline but ensures you have the appropriate data needed for each step of the procress. Similarly, explicit data definitions could also make things easier to test because you can write tests using synthetic data or even generate random data for Monte Carlo simulations. This way, your IDE's static analyzer can take advantage of intellisense or other autocomplete tools to help you code faster and with many errors caught prior to even running your code.
More concretely, I recommend creating data encapsulation classes that (1) use factory methods for class creation instead of constructors; (2) are immutable, or cannot be changed after creation; (3) include some level of data validation. In the following sections I will be drawing on the examples in this notebook for illustration.
Class factory methods can be used to handle the mappings between your source data (e.g. json or csv formatted data) and the object instances. Among other benefits, they are especially convenient in cases where your input data may come from multiple source formats. For instance, say your input data is split into two files with different formats but essentially correspond to the same type of data elements. You could create two separate factory methods - one to handle each. This also makes it possible to create your classes immutable. This approach follows a principle from the Zen of Python: "in the face of ambiguity, refuse the temptation to guess."
To make these points clearer, I will make an example out of the iris dataset we have been looking at so far. Dataclasses have been extremely popular lately as they provide convenient ways to create classes meant for storing data. The example below shows my example dataclass with three properties and two class factory methods - one for creating the object from a dictionary, and one for creating the object from a dataframe row.
import dataclasses
@dataclasses.dataclass(frozen=True)
class IrisEntryDataclass:
sepal_length: int
sepal_width: int
species: str
# factory method constructor
@classmethod
def from_dataframe_row(cls, row: pd.Series):
new_obj: cls = cls(
sepal_length = row['sepal_length'],
sepal_width = row['sepal_width'],
species = row['species'],
)
return new_obj
@classmethod
def from_dict(cls, data: dict):
new_obj: cls = cls(
sepal_length = data['sepal_length'],
sepal_width = data['sepal_width'],
species = data['species'],
)
return new_obj
We can simply iterate through each row of the dataframe and use the factory method to convert the dataframe rows to these dataclasses.
entries = list()
for ind, row in iris_df.iterrows():
new_iris = IrisEntryDataclass.from_dataframe_row(row)
entries.append(new_iris)
Alternatively we could create the objects from the parsed json data we showed in the previous example about nested iterables. This line of code produces the exact same result.
iris_dict = iris_df.to_dict(orient='records')
entries = [IrisEntryDataclass.from_dataframe_row(d) for d in iris_dict]
entries[:3]
Output:
[IrisEntryDataclass(sepal_length=5.1, sepal_width=3.5, species='setosa'),
IrisEntryDataclass(sepal_length=4.9, sepal_width=3.0, species='setosa'),
IrisEntryDataclass(sepal_length=4.7, sepal_width=3.2, species='setosa')]
The advantage is that Python and our static analyzers providing autocomplete and error analysis know that these are all valid properties of the object. If we ran this code:
entries[0].some_property
Our IDE would give us a warning that objects of type IrisEntryDataclass have no property called some_property, and we cannot assign a value to it because we used frozen=True - our class is immutable. This provides strict enforcement of our data format - we know for sure that each instance of this class will have exactly the specified properties, and no more. If we have a function that accepts a list of IrisEntryDataclass objects, we (and our IDE) know exactly which properties are available to us.
Consider using attrs
While dataclasses are useful, I recommend going a step further by using attrs to include explicit data validation. The attrs package provides some convenient methods for creating classes meant for storing data, so that may be preferable in many cases. Note: attrs provides a superset of the features found in dataclasses because it acted partially as their inspiration for inclusion in the standard library.
See: Example Notebook
I will not include my implementation of the attrs version of the same class, but instead direct you to Example 2 of my example notebook. In that class I use the frozen=True and slots=True settings to make sure the object is immutable (cannot be changed) and uses slots (see this explanation), which reduces memory usage and prevents the user from creating attributes later on.
I implemented validation using several methods. I used the converter parameter of attrs.field() to enforce data conversion - in addition to throwing errors for non-convertable types, this can prevent your code from being sensitive to the way that the data was ingested (e.g. whether pandas converted a column to a string). I also added two data validation methods using decorators recognized by attrs: species_validator and meas_validator; these define the criteria of the data being inserted upon instantiation. Note that because I did not use the default parameter in attrs.field, all of this data must be provided during instantiation. This is my class definition:
This class provides gaurantees about the structure of your data - if your class method constructor runs without errors, you know that your data structure will have the required data early in your pipeline. Furthermore, all validation and data structuring is done as part of the object - you won't need to write an additional validation script to verify that the data meets some assumed criteria. Multiple methods for producing the same dataset can be implemented in the same class. In this way, all checks and gaurantees about your data exists in this single class. If you want to change assumptions or structure of the data, it will appear here.
Wrap collections
I also recommend using class objects for higher-order data structures composed of multiple pieces of data or even data in different formats. While this can be dangerous in general, in certain select scenarios I recommend creating collections classes that inherit directly from builtin iterable types. This can eliminate a lot of the boilerplate __iter__, __getitem__, and __len__ code that we often need when encapsulating iterables, and also allows us to wrap factory constructor methods that produce multiple data objects - see the factory method from_dataframe here. I also implemented a couple useful functions for working with lists of this specific data type. Functions for grouping and filtering these objects also fit better here than in downstream consumers.
class IrisEntriesList(typing.List[IrisEntryAttrs]):
######################### Factory Methods #########################
@classmethod
def from_dataframe(cls, df: pd.DataFrame):
# add type hint by hinting at returned variable
elist = [IrisEntryAttrs.from_dataframe_row(row) for ind,row in df.iterrows()]
new_entries: cls = cls(elist)
return new_entries
######################### Grouping and Filtering #########################
def group_by_species(self) -> typing.Dict[str, IrisEntriesList]:
groups = dict()
for e in self:
groups.setdefault(e.species, self.__class__())
groups[e.species].append(e)
return groups
def filter_sepal_area(self, sepal_area: float):
elist = [e for e in self if e.sepal_area() >= sepal_area]
entries: self.__class__ = self.__class__(elist)
return entries
By always using factory methods here to create the sequence of IrisEntries, we are creating further gaurantees for the structure of the data inside these objects. You could accomplish the same thing by inheriting through composition - and indeed in many cases this is safer - but direct iterable inheritance is better than nothing, in my opinion.
In my experience, adding more structure to your data pipelines - at least where the actual data is concerned - can improve the quality of your code and the speed with which you produce results, even if it requires more time and dicipline up-front. While speed is important in producing results, nothing is more important than getting the analysis right.
2. Version almost everything
My next recommendation is related to managing projects. A key challenge in project management is keeping track of different intermediary data products and the code used to generate them. It is typical to be in a scenario where, halfway through building a data pipeline, your requirements change or you produce results that encourage you to take a different approach to analysis. In that case, you don't want to delete the old intermediary results because you may want to refer back to them later. Or, worse yet, you shared them with a client and may need to refer back to them for their sake. Either way, you also want to keep track of the code you used to create those files, in case you get any questions about it or are simply curious. It is in this scenario that I recommend that you retain multiple class definitions for the same type of object and keep track using naming conventions. Then, when you go to look closer at a result you generated previously, you can easily track the code you used to generate it.
It might be worth thinking more about typical data science workflows to get a sense of why this might make sense. First, lets take a look at the following figure.
At the top of this figure we see the original dataset, composed of one csv file and one json file. Each of the scripts that appear here involve some type of transformation from one data type to another. Script 1 takes the original csv data and converts it into a new Intermediate csv file. Script 3 takes the orignal csv file, the original json file, and the intermediate csv file generated by Script 1, and converts it into Final table csv file. Script 4 then converts this table into a visualization that will be presented to the client. Note that on the right we also see that Script 2 converts the original json data to Some figure png file - possibly for another analysis.
In an ideal world, we work on and finish Script 1, then work on and finish Script 2, and so on until we finish coding and can produce the final data from the original data. In a more practical scenario, most likely will need to go back and fix some upstream script to add or edit some information according to requirement changes or just changing realization of the information needed to produce intermediary results. When we do this, however, it doesn't always make sense to delete data that was previously generated - either so we can use it as reference or present it as intermediary results to the client. In these cases, it makes the most sense to keep track of the data and the code that was used to generate it through an informal versioning system of your design.
For an object-oriented example, let's assume that the client gives you data in the format represented by the class IrisA.
@dataclasses.dataclass
class IrisA:
sepal_length: int
sepal_width: int
species: str
You write a script that converts IrisA data to IrisB data using its class method constructor from_a, and you saved the result as resultB.csv. This result will be used later on by another script to produce the final figure. Lets say you produce the final figure and call it finalB.png, then share it with your client as intermediate results. See my example class below.
@dataclasses.dataclass
class IrisB:
sepal_area: float
@classmethod
def from_a(cls, a: IrisA):
return cls(sepal_area = a.sepal_length * a.sepal_width)
The client has some suggested improvements, and you realize you need one additional piece of information from the IrisA dataset to be included in IrisB: the species. So you go back and edit IrisB to include this information. You use this dataset to produce the final result finalB2.png, and share with the client.
@dataclasses.dataclass
class IrisB:
sepal_area: float
species: str
@classmethod
def from_a(cls, a: IrisA):
return cls(sepal_area = a.sepal_length * a.sepal_width, species=a.species)
The client asks about a discrepency between finalB.png and finalB2.png - can you answer their question? You could go back to the correct commit based on datetimes, but that might be a little tricky if the file timestamp was changed after copying/etc. Instead, I recommend creating a versioning system that is consistent between the names of your data objects and the intermediate data files they were used to generate.
@dataclasses.dataclass
class IrisB_v1:
sepal_area: float
@classmethod
def from_a(cls, a: IrisA):
return cls(sepal_area = a.sepal_length * a.sepal_width)
@dataclasses.dataclass
class IrisB_v2:
sepal_area: float
species: str
@classmethod
def from_a(cls, a: IrisA):
return cls(sepal_area = a.sepal_length * a.sepal_width, species=a.species)
When you save the files, you could integrate the name of the class into the filename. For instance, you could call them result_IrisB_v1.csv and result_IrisB_v2.csv, which you could then translate to a final figure names like final_result_IrisB_v1.png final_result_IrisB_v2.png. This is what it might look like:
a = IrisA(1.0, 2.0, 'big')
b1 = IrisB_v1.from_a(a)
save(b1, f'result_{b1.__class__}.csv')
b2 = IrisB_v2.from_a(a)
save(b2, f'result_{b2.__class__}.csv')
Of course every case is unique and I don't recommend using this everywhere, but I have found it to be useful in scenarios where I am getting a lot of client feedback and I need to refer to old code when I'm trying to explain differences between results figures or intermediate data.
3. Create parameter or settings objects that live in your code
Building off of the previous two examples, I recommend creating parameter objects that you can use to keep track of results created with different parameters. This essentially follows from the previous two examples, but further introduces the possibility that you may generate results from many different parameter sets over the lifetime of your project. Ideally the defined parameter set will be used at every step of your data pipeline to make it perfectly reproducable. Lets see the example below.
Start by creating a dataclass to contain parameter information. This defines all the variables that will be used to generate results throughout your data pipeline. It might be helpful to have some version_name member to make it easy to get the version as a string. This even includes a parameter that accepts a type - this will allow you to control which version of a class is being used throughout your pipeline. This may be overkill, but you can imagine projects that are of sufficient complexity as to benefit to this level of detail.
@dataclasses.dataclass
class Params:
version_name: str
IrisB: type
intermediate_fname: str
final_fname: str
Now I recommend creating functions or module-level variables for param object instances, including the version number as part of the object name. Here I have two params objecs, which I named 0x1 and 0x2.
def params_0x1() -> Params:
return Params(
version_name = '0x1',
IrisB = IrisB_v1,
intermediate_fname = 'intermediate_0x1.csv',
final_fname = 'final_0x1.csv',
)
def params_0x2() -> Params:
return Params(
version_name = '0x2',
IrisB = IrisB_v2,
intermediate_fname = 'intermediate_0x2.csv',
final_fname = 'final_0x2.csv',
)
So at the beginning of each script you can instantiate a params object and use it at various points.
params = params_0x1()
a = IrisA(1.0, 2.0, 'big')
b = params.IrisB.from_a(a)
save(b, f'result_{params.version_name}.csv')
This approach may be overkill in some scenarios, but if the project gets large enough it is an option you may want to consider.
4. Make I/O explicit in your top-level scripts
This principle is very simple: make it such the user can see save and read functions at the top level of your script (even if the filenames are hidden). It should be clear to the reader that the script is ingesting one or more datasets and exporting others. Adding this to your script can save you a lot of time later when you try to determine which types of data this script makes and outputs.
df = pd.read_csv('test.csv')
new_data = myfunc(df)
new_data.save_json('new_data.json')
This can still be consistent with previous recommendations as long as you access filename properties at the top level of your script. In this example, the filenames are stored as members of the params object.
params = params_0x1()
df = pd.read_csv(params.test_csv_fname)
new_data = myfunc(df)
new_data.save_json(params.output_json_fname)
5. Include validation when possible
Validation has the potential to drastically increase your speed as an analyst because it allows you to build assumptions about your data into your production pipeline directly. As you begin to build your pipeline, you will start to ask questions about your dataset that you can typically answer through exploratory analysis. This requires overhead, however, because you must move back-and-forth between the exploratory analysis and the pipeline code. Furthermore, if your data changes or you add additional data, you need to remember to re-run your exploratory analysis code and verify that the same assumptions hold. Instead, I recommend that you make assumptions as you build your pipeline and integrate them as part of your production code (to be disabled later if needed). You can always change these assumptions as you learn more about your data, but I have found that it is best to implement them early in development.
I typically recommend implementing validation as far upstream as possible - ideally when the data is ingested into an object - often this means within constructors or factory methods. When possible, this is easier than validating results downstream after some transformations have happened. For instance, it would be better to validate that an age value is greater or equal to zero within a class constructor than it is to check every value before calculating an average. Also consider inserting validation code during the exploratory phase of your analysis, when you are implicitly validating assumptions anyways.
Of course, validation is also helpful for detecting issues in your code. While unit tests tend to be a gold standard, you may not have the time or desire to implement a full-featured test suite. In lew of that, I at least recommend you include exceptions or assertions at various points in your pipeline - especially when they are obvious or come before a portion of code that might be particularly difficult to debug if your assumptions do not hold.
Validation in Python
In Python, there are generally two options for including validation in your production pipeline: assertions and exceptions. Use assertions when you want a full-stop on your program if an assumption does not hold. You may want to use assertions if there is a major error upstream that will cause any subsequent results to be nonsense. In this case you will likely need to go back into full development mode to fix the issues that arose before you can begin to run anything. Assertions may also be the best option if you are lazy. Alternatively, if there are cases in which you may want to ignore particular assumptions (at least momentarily), you can raise exceptions when an assumption in your data does not hold. It is generally best to create custom exceptions to handle these issues so that when you go to ignore or do some logic based on the error, you are catching the correct issue.
In Python you will ideally create custom exceptions for your applications - a process which is easier than you might think. Now I will illustrate how you can use exceptions using an example class called Person created using attrs with some validator functions. The object has two properties: age and full_name. There are several things that can be wrong with a Person entry, so we will later add validators with custom exceptions to indicate which aspect of the data is wrong. We start by making the custom exceptions. First, the most obvious issue would be that the age is too high or below zero - the entry is obviously wrong if that is the case. So, we make a custom exception that inherits from ValueError (we could also inherit from BaseException, but this has some utility).
class PersonAgeOutOfRangeError(ValueError):
pass
Next we want to raise an exception if there is an issue with the full_name of the Person. This is an interesting case because two things can be wrong: full_name can be an empty string or it can only include a single word when at least two words are needed to make a full name (at least in most of the Americas). Because they are both issues with the name, we can start by making an exception PersonNameEror to be raised when the name is out of range. But we can also improve granularity of this exception by indicating whether the issue was that the name string was empty or whether they only had one name. For that, we create two additional exceptions - one for empty name, and the other for a last name error.
class PersonNameEror(ValueError):
pass
class PersonNameWasEmptyError(PersonNameEror):
pass
class PersonFirstLastNameError(PersonNameEror):
pass
Now we can define the Person class with validation functions using attrs. Notice that validate_age and validate_name are both added to the __init__ constructor generated by the attr.s decorator.
@attr.s
class Person:
age: int = attrs.field(converter=float)
full_name: int = attrs.field(converter=str)
@age.validator
def validate_age(self, attr, value):
if value < 0 or value > 150:
raise PersonAgeOutOfRangeError(f'{attr.name} must be between '
f'0 and 150')
@full_name.validator
def validate_name(self, attr, full_name: str):
if not len(full_name):
raise PersonNameWasEmptyError(f'{attr.name} cannot '
f'be empty string.')
elif not len(full_name.split()) > 1:
raise PersonFirstLastNameError(f'{attr.name} requires '
f'first and last names.')
Instantiating this object is simple. When we create this person, we get no errors.
Person(age=10, full_name='yo holla')
However, we do get an error when we provide an incorrect age:
Person(age=-1, full_name='jose dillan')
Exception raised:
__main__.PersonAgeOutOfRangeError: age must be between 0 and 150
Now try making a person with a single name:
Person(age=10, full_name='Jose')
Exception raised:
__main__.PersonFirstLastNameError: full_name requires first and last names.
This is great - if these assumptions are violated, we will see an error in our code and we can decide how to handle it. Hypothetically, let's say you received full names and ages of some individuals from a survey. As we know, survey data is messy; in this case, some respondents did not provide their full names, but we need the full names for our analysis. In this case, we still want to make assumptions about age and name cannot be an empty string - the program should fail if those are violated. We would, however, simply like to drop the entries where the full name was not provided. To do this, we can use try and catch keywords to implement exception handling.
We start with a dataset that looks like this:
person_data = [
{'age': 22, 'name': 'jose rodriguez'},
{'age': 5, 'name': 'johnny'},
{'age': 30, 'name': 'carol carn'},
{'age': 55, 'name': 'david duke'},
]
And we want to make a list of Person objects from this dataset. To do this, we can wrap the person constructor in a try statement and choose to ignore the case when the PersonFirstLastNameError exception was raised. In this way, entries that don't have first and last names are dropped from the list and will not be included in the dataset. The program will still crash if one of the other two exceptions was raised, however. Using this technique, we can choose to ignore entries with one type of error while still retaining the other two assumptions.
people = list()
num_invalid = 0
for d in person_data:
try:
people.append(Person(age=d['age'], full_name=d['name']))
except PersonFirstLastNameError:
num_invalid += 1
If, however, we wanted to ignore persons with any type of name issue, we can either catch multiple exceptions or use the more generic exception PersonNameEror. Here we catch any exception which is a subclass of PersonNameEror.
try:
Person(age=pd['age'], full_name=pd['name'])
except PersonNameEror:
pass
We can also capture multiple exceptions at once - useful if you want to integrate more validation checks in the future but do not necessarily want to drop those rows.
try:
Person(age=d['age'], full_name=d['name'])
except (PersonFirstLastNameError, PersonNameWasEmptyError):
pass
If, however, you want to relax any of the validation functions so they don't raise exceptions, you'll need to update your validation logic.
Including validation for assumptions about the data and pipeline can add additional gaurantees about your data that will be useful later. Through use of exceptions and assertions in validators, you will know that every instance of your object will maintain a set of characteristics that your analyses are predicated on - and, as the analyst, you can make intentional calls about which errors to ignore and how to handle errors that do arise.